Comprehensive online database deployed for exploring massive public plant RNA-seq data
Chris Edwards 2020-08-15
Applications of Next Generating Sequencing (NGS) technology in transcriptome profiling have greatly improved our understanding of transcriptional regulation at a genome-wide scale in the last decade. The research community has produced tens of thousands of RNA-sequencing (RNA-seq) libraries. However, accessing such a huge amount of RNA-seq data poses a big challenge for groups that lack dedicated bioinformatic personnel or expensive computational resources.
On August 4, Associate Professor Jixian Zhai (Institute of Plant and Food Science, Department of Biology) led his research team to publish a paper in the high-impact academic journal Molecular Plant (IF = 12.084). The paper was titled “A comprehensive online database for exploring ~20,000 public Arabidopsis RNA-Seq libraries”.
The Arabidopsis RNA-seq database (ARS, http://ipf.sustech.edu.cn/pub/athrna/) is a “Google-style” website that integrates 20,068 publicly available Arabidopsis RNA-seq library data deposited at GEO, SRA, ENA, and DDBJ. ARS calculated the gene expression level at each library and performed co-expression analysis. ARS also classified all libraries into 1176, 1102, 12, and 176 groups of mutants, treatment conditions, tissues and development stages, respectively, and analyzed the differentially expressed levels of all genes under different mutants or treatments.
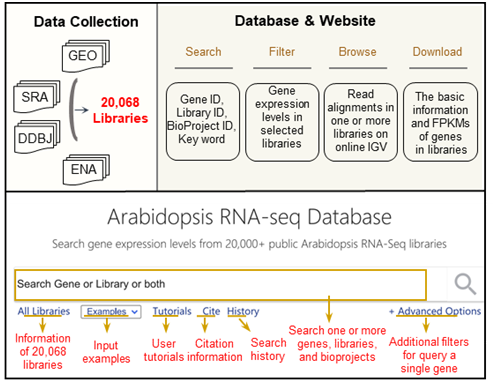 Figure 1: Overview of the construction of Arabidopsis RNA-Seq Database (ARS).
Figure 1: Overview of the construction of Arabidopsis RNA-Seq Database (ARS).
ARS supports queries of gene IDs, library IDs, BioProject IDs, keywords, or a combination of these query types to show expression levels of specific genes in selected libraries in the form of one or more tables and diagrams. A built-in online Integrative Genomics Viewer (IGV) supports viewing detailed read alignments of each library, and the users can search and select libraries by keywords. The search results can be easily shared with others by clicking the “Share” button to generate a link for the current page.
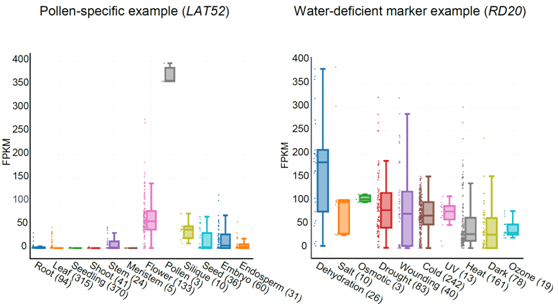
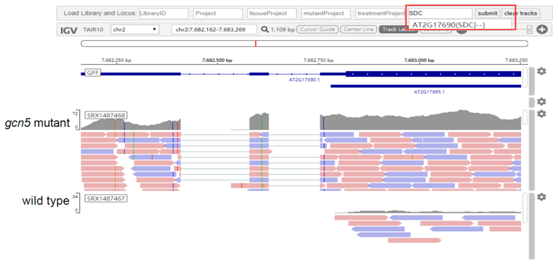 Figure 2: The expression levels among different tissues and stress (top), an overview of Online IGV (bottom).
Figure 2: The expression levels among different tissues and stress (top), an overview of Online IGV (bottom).
Associate Professor Jixian Zhai is the corresponding author of the paper. The co-first authors are Research Assistants Hong Zhang & Fei Zhang, and Ph.D. student Yiming Yu. This paper is supported by the National Natural Science Foundation of China, the Program for Guangdong Introducing Innovative and Entrepreneurial Teams, and the Shenzhen Sci-Tech Fund.