Researchers make significant progress in high-throughput drug target screening technology
2023-09-12
With the development of molecular biology techniques and the completion of the Human Genome Project, numerous genes are unraveled as potential targets for drug discovery. However, not all of these genes are relevant to the disease development process. Selecting and validating targets in the process of new drug development is crucial and serves as the foundation and assurance for drug innovation. Despite the rapid progress in understanding the biological mechanisms underlying diseases, the discovery of new drug targets remains an expensive and time-consuming process. On the other hand, recent works have revealed many existing drugs actually work through unknown off-targets that open up new strategies to expedite target and drug discovery.
To address the aforementioned challenges, a research team led by Associate Professor Chris Soon Heng Tan from the Department of Chemistry at the Southern University of Science and Technology (SUSTech) has developed a high-throughput mixing strategy called Matrix-Augmented Pooling Strategy (MAPS). Their study focused on the research of cell-specific drug-target interactions using high-throughput drug target screening technology. The team proposed a high-throughput mixing strategy, MAPS, to identify drug targets. This method was used to analyze the interactions between drugs and targets in five different cell lines.
Their research work, entitled “Target deconvolution with matrix-augmented pooling strategy reveals cell-specific drug-protein interactions”, has been published in the academic journal Cell Chemical Biology. The results revealed differences in target interactions and binding affinities between different cell lines. Furthermore, the study validated that BRAF and CSNK2A2 are potential off-targets for the drugs bafetinib and abemaciclib, respectively.
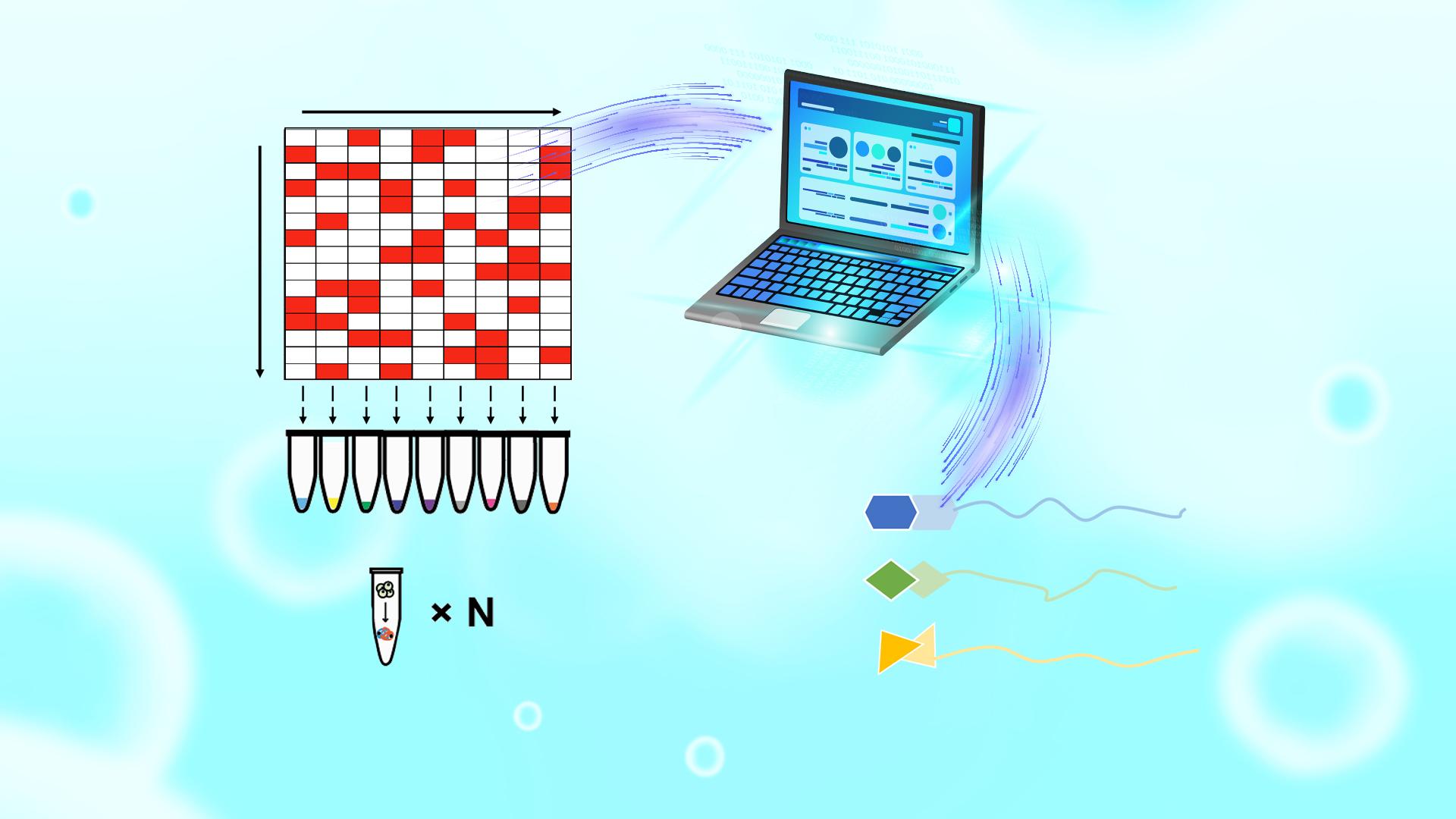
This strategy used by the researchers involved optimizing the arrangement of multiple drugs in mixtures and mathematically processing the mixtures to simultaneously depict the targets of each drug. The technique consists of three main steps (Figure 1): 1) optimizing the arrangement of drugs to ensure high orthogonality between groups, with similar or equal numbers of compounds in each group and each compound present in at least three groups; 2) creating multiple mixture systems of the test drugs based on the arrangement matrix and using iTSA single-temperature point heating to obtain protein thermal stability transition results; 3) analyzing the mass spectrometry results to identify the drug targets.
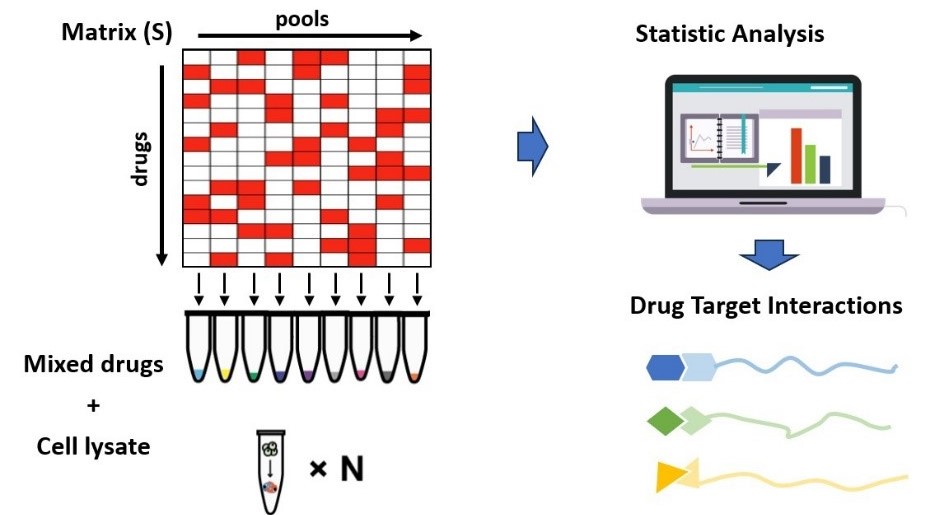
Figure 1. Schematic of the MAPS workflow.
The researchers validated this strategy through Thermal Proteome Profiling (TPP) tests on 15 different drugs. This approach increased experimental throughput by 60 times and reduced costs by 90%, offering a promising method for large-scale drug target screening. By conducting the same set of MAPS experiments on five cell lines – K562, MCF7, HEK293T, HCT116, and HepG2 – including five potential off-target drugs with ambiguous mechanisms of action, MAPS-iTSA analysis revealed varying interaction affinities between drugs and target proteins across different cell lines (Figure 2). Data analysis from different cell lines identified numerous new potential off-targets for these drugs.
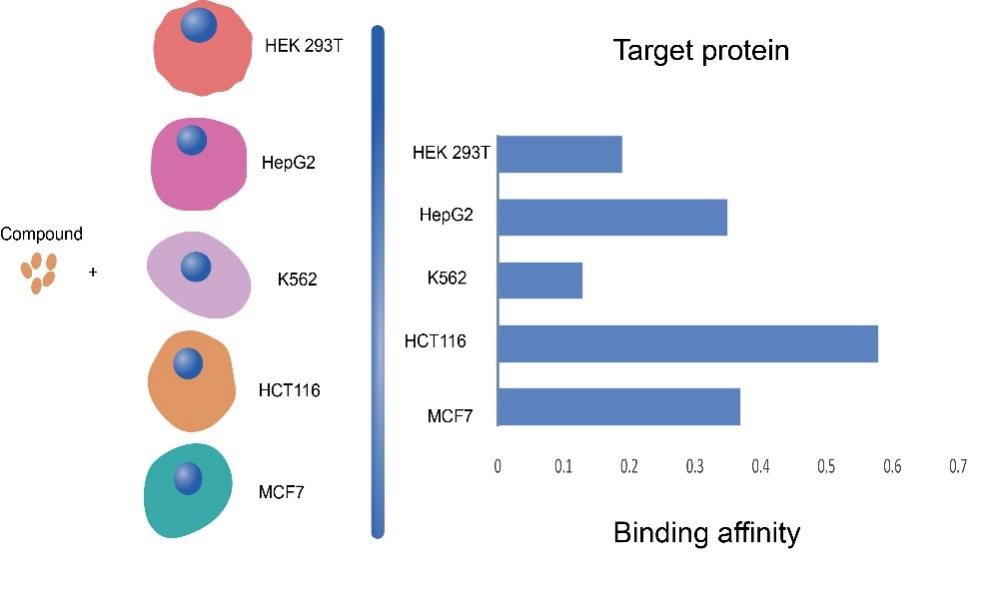
Figure 2. Differential affinities of drug-target interactions in different cell lines.
Analysis showed that Bafetinib, an experimental agent targeting BCR-ABL and LYN for lymphocytic leukemia, binds BRAF. Bafetinib-BRAF affinity varies between cell lines; as shown in Figure 3, the potential targeting of BRAF by bafetinib was validated through ITDRFCETSA (isothermal dose-response), NanoBRET (intracellular kinase detection), and Kinase Hotspot Assay (radioisotope-labeled ATP method). In addition, the researchers also confirmed that CSNK2A2 is a potential target for abemaciclib, a drug that supposedly targets CDK4/6 and is used in the treatment of breast cancers. This strategy offers a robust approach for analyzing drug-target affinities in different cell types and enables large-scale drug target screening.
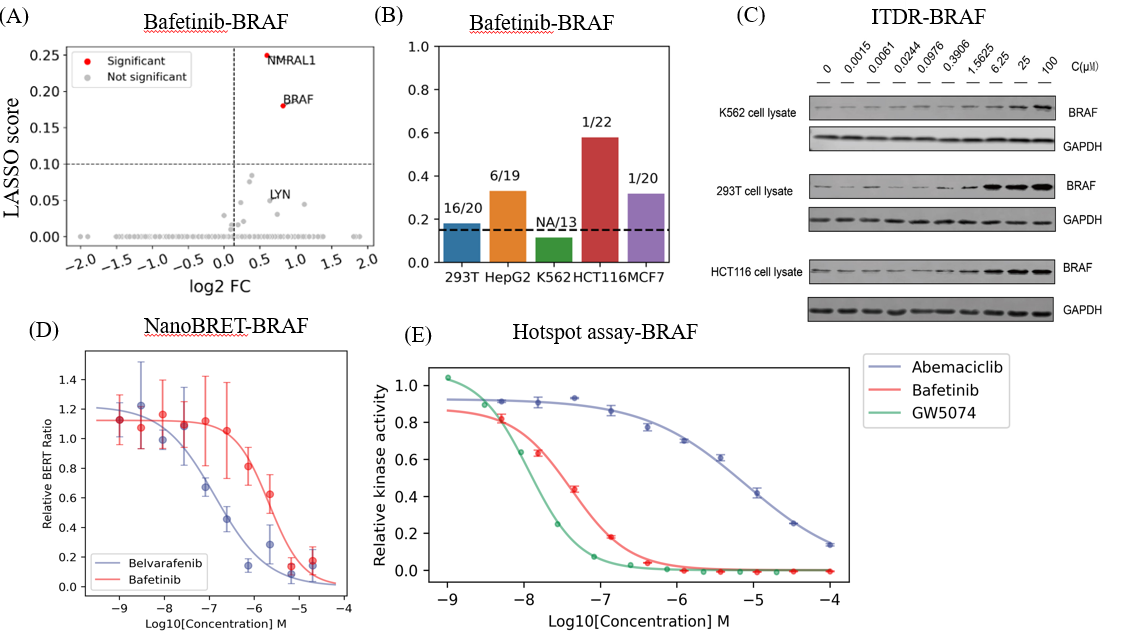
Figure 3. Off-target explanation and validation. A) Target volcano map of bafitinib drug. B) Affinity of Bafetinib-BRAF in different cell lines. C) Western blot of dose-dependent CETSA of bafetinib-BRAF. The changing of blots is consistent with the LASSO score of MAPS-iTSA. D) Dose-response curve for NanoBERT in vivo off-target validation of bafetinib-BRAF. E) Dose-response curve for kinase hotpots in-vitro off-target validation of bafetinib-BRAF.
In summary, the research led by Chris Soon Heng Tan’s team at SUSTech has achieved important advancements in high-throughput drug-target screening technology, particularly in studying cell-specific drug-target interactions. Their innovative MAPS method offers a valuable approach to identifying drug targets and has potential applications in new drug development.
Hongchao Ji, a postdoctoral researcher from the Department of Chemistry at SUSTech and currently an Associate Researcher of the Agricultural Genomics Institute at Shenzhen (AGIS) by the Chinese Academy of Agricultural Sciences (CAAS), along with Xue Lu, a Ph.D. candidate from SUSTech, are co-first authors of this paper. Associate Professor Chris Soon Heng Tan from SUSTech is the first corresponding author, and SUSTech is the first and corresponding communication unit. Collaborating institutions include the University of California and the Nuffield Department of Medicine at the University of Oxford.
This research was supported by the Shenzhen Science and Innovation Commission and the National Natural Science Foundation of China (NSFC).