SUSTech researchers develop novel technique to understand RNA processing in plants
2020-07-09
New research conducted at Southern University of Science and Technology (SUSTech) may have determined the kinetics of splicing in plants using Nanopore-based sequencing. It could lead to a better understanding of the RNA splicing process in plants.

On June 15, Associate Professor Jixian Zhai (Institute of Plant and Food Science, Department of Biology) led his research team to publish a ground-breaking paper in the high impact academic journal Nature Plants (IF = 13.297). The paper was titled “Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants.”
Intron splicing is a crucial step in eukaryotic mRNA maturation. It is generally considered to occur co-transcriptionally. In yeast, half of the introns are already spliced when RNA polymerase-II (Pol-II) is only 45 nt downstream of introns.
On the other hand, the splicing kinetics, the link between splicing and transcription, and the relationship between splicing and polyadenylation remain mostly unknown, particularly in multicellular organisms like plants.
The researchers developed a Nanopore-based full-length sequencing method to profile chromatin-bound nascent RNA. They found that more than half of introns in the model plant Arabidopsis thaliana were transcribed 1 kb downstream from Pol-II had not been cut yet. That reflects a much slower speed than yeast.
They also found that many of the full-length chromatin-bound RNA molecules are polyadenylated, while still containing unspliced introns at specific positions. These introns are nearly absent in the cytoplasm. The introns are also resistant to nonsense-mediated decay, suggesting that they are post-transcriptionally spliced before the transcripts are released into the cytoplasm, and thus are termed post-transcriptionally spliced introns (pts-introns).
Analysis of around 6,500 public RNA-sequencing libraries revealed that the splicing of pts-introns requires the function of splicing-related proteins such as PRMT5 and SKIP, and is also influenced by various environmental signals. The majority of the intron retention events in Arabidopsis are at pts-introns. It suggests that chromatin-tethered post-transcriptional splicing is a significant contributor to the widespread intron retention that is observed in plants, and could be a mechanism to produce fully spliced functional mRNAs for rapid response.
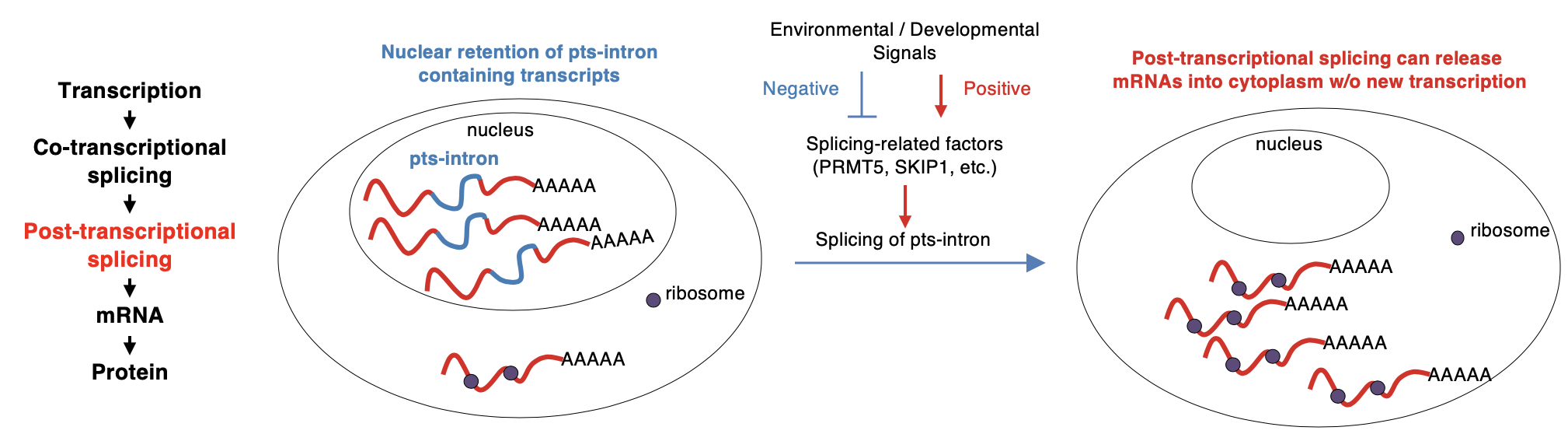 Figure 1: A model for nuclear retention and post-transcriptional splicing as an important layer of post-transcriptional regulation
Figure 1: A model for nuclear retention and post-transcriptional splicing as an important layer of post-transcriptional regulation
The Nanopore-based chromatin-bound RNA sequencing method developed by the team can capture full-length nascent transcripts. These transcripts include elongating transcripts, read-through transcripts, and polyadenylated transcripts. The method enables the simultaneous detection of splicing status, Pol-II transcription position, and polyadenylation at the genome-wide scale. It provides an essential tool for researchers to track and study the dynamic processes of mRNA transcription, splicing, processing, and maturation.
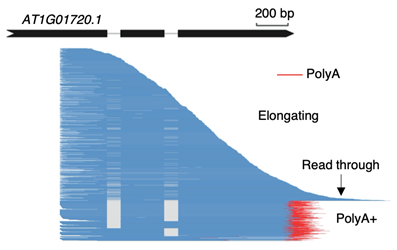 Figure 2: An example of full-length nascent transcripts captured by Nanopore-based chromatin-bind RNA sequencing method
Figure 2: An example of full-length nascent transcripts captured by Nanopore-based chromatin-bind RNA sequencing method
Associate Professor Jixian Zhai is the corresponding author of the paper. The co-first authors are research assistant professors Dr. Jinbu Jia and Dr. Yanping Long. Additional contributions came from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences, and South China Agricultural University.
The research paper received support from the National Key R&D Program of China, the National Natural Science Foundation of China (NSFC), the Program for Guangdong Introducing Innovative and Entrepreneurial Teams, and the Shenzhen Sci-Tech Fund, the China Postdoctoral Science Foundation, the Chinese Academy of Sciences (Strategic Priority Research Program), the Youth Innovation Promotion Association of CAS, and the State Key Laboratory of Plant Genomics.